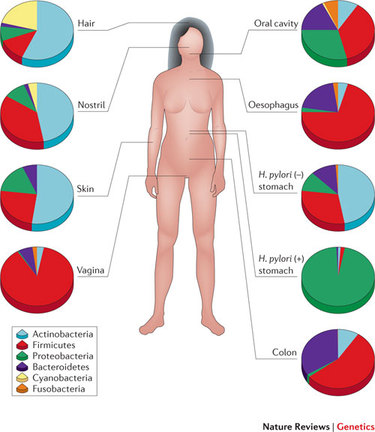
 * Image from Cho and Blaser (2012) Nature Reviews Genetics
* Image from Cho and Blaser (2012) Nature Reviews Genetics A catalogue of microbes in the human gut
It comes to my attention how our knowledge of the microbial world is rapidly expanding throughout the years. As of today, researchers are able to identify most of the microorganisms that inhabit a specific environment in an ecosystem, including parts of our bodies (see image on the left). Now is known, that the set of microbes, their genomes and their environmental interactions that are associated within our bodies (here forth termed as the human microbiome) have an important role in human health. This is reason of why many scientists around the world are paying attention to microbes and their role in diseases and other medical-related conditions.
The human microbiome project was developed with the purpose of understanding all the microbial communities (including their genomes and their interactions) that are associated to humans. Their aim, as is presented on their website, reads as follow:
It comes to my attention how our knowledge of the microbial world is rapidly expanding throughout the years. As of today, researchers are able to identify most of the microorganisms that inhabit a specific environment in an ecosystem, including parts of our bodies (see image on the left). Now is known, that the set of microbes, their genomes and their environmental interactions that are associated within our bodies (here forth termed as the human microbiome) have an important role in human health. This is reason of why many scientists around the world are paying attention to microbes and their role in diseases and other medical-related conditions.
The human microbiome project was developed with the purpose of understanding all the microbial communities (including their genomes and their interactions) that are associated to humans. Their aim, as is presented on their website, reads as follow:
"The aim of the HMP is to characterize microbial communities found at multiple human body sites and to look for correlations between changes in the microbiome and human health."
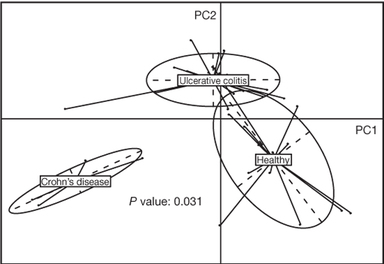 *Figure from Qin, et al. (2010) Nature
*Figure from Qin, et al. (2010) Nature In addition to the human microbiome project, researchers from Europe and China collaborated to develop the MetaHit (Metagenomics of the Human Intestinal Tract) project. This project lasted from 2008 – 2012, and with it, many scientific discoveries were published (look video below for more information about the project). Among those discoveries, it is worthwhile to mention the work of Qin, et al. (2010) from the MetaHIT Consortium, about the human gut microbial gene catalogue.
In this article, using deep metagenomic sequencing, the authors managed to identify and characterize a catalogue of microbial genes (encoded in their metagenomes) that were present in the gut of individuals with obesity/diabetes phenotypes, patients with Crohn's disease or ulcerative colitis, and healthy individuals. They were able to study these metagenomes due to the power of the new emerging sequencing technologies. Their results showed that the microorganisms that predominated in the gut of the individuals that were studied belong to the phyla Firmicutes and Bacteroidetes. In addition, they noted a significant difference in the species abundance of the core microbiota of healthy individuals and patients with Crohn's disease or ulcerative colitis (figure shown above). These results might suggest a possible role of specific microbial species in the progression of inflammatory bowel disease in patients suffering from such diseases.
With the advancement of these sequencing technologies, we could easily predict how medicine will change in the near future. With years to come, and as long as technologies continue to grow, we might be able to diagnose and medicate patients individually according to their genomic and metagenomic background. In this way, treatments could be more specific and more accurate, leading to a better diagnosis and a faster recovery.
*References:
(1) Cho, I., and Blaser, M.J (2012) The human microbiome: at the interface of health and disease. Nature Reviews Genetics 13: 260–270.
(2) Qin, J., Li, R., Raes, j., et al. (2010) A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464(4): 59–67.
In this article, using deep metagenomic sequencing, the authors managed to identify and characterize a catalogue of microbial genes (encoded in their metagenomes) that were present in the gut of individuals with obesity/diabetes phenotypes, patients with Crohn's disease or ulcerative colitis, and healthy individuals. They were able to study these metagenomes due to the power of the new emerging sequencing technologies. Their results showed that the microorganisms that predominated in the gut of the individuals that were studied belong to the phyla Firmicutes and Bacteroidetes. In addition, they noted a significant difference in the species abundance of the core microbiota of healthy individuals and patients with Crohn's disease or ulcerative colitis (figure shown above). These results might suggest a possible role of specific microbial species in the progression of inflammatory bowel disease in patients suffering from such diseases.
With the advancement of these sequencing technologies, we could easily predict how medicine will change in the near future. With years to come, and as long as technologies continue to grow, we might be able to diagnose and medicate patients individually according to their genomic and metagenomic background. In this way, treatments could be more specific and more accurate, leading to a better diagnosis and a faster recovery.
*References:
(1) Cho, I., and Blaser, M.J (2012) The human microbiome: at the interface of health and disease. Nature Reviews Genetics 13: 260–270.
(2) Qin, J., Li, R., Raes, j., et al. (2010) A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464(4): 59–67.
 RSS Feed
RSS Feed